
Opportunities
We are currently seeking:
Applicants for scholarships for 3- to 6-month exchanges to Norwegian master programs at the University of Oslo (UiO), the University of Bergen (UiO), or the Norwegian University of Science and Technology (NTNU). Exchanges to start in 2025. Click here for more information.
Potential master projects
Below you can welcome to explore some exciting ideas for master projects that BiGTREE students can develop during research stays abroad. Contact us for more information about any of these possibilities.
Hosted by UNMSM (Peru)
-
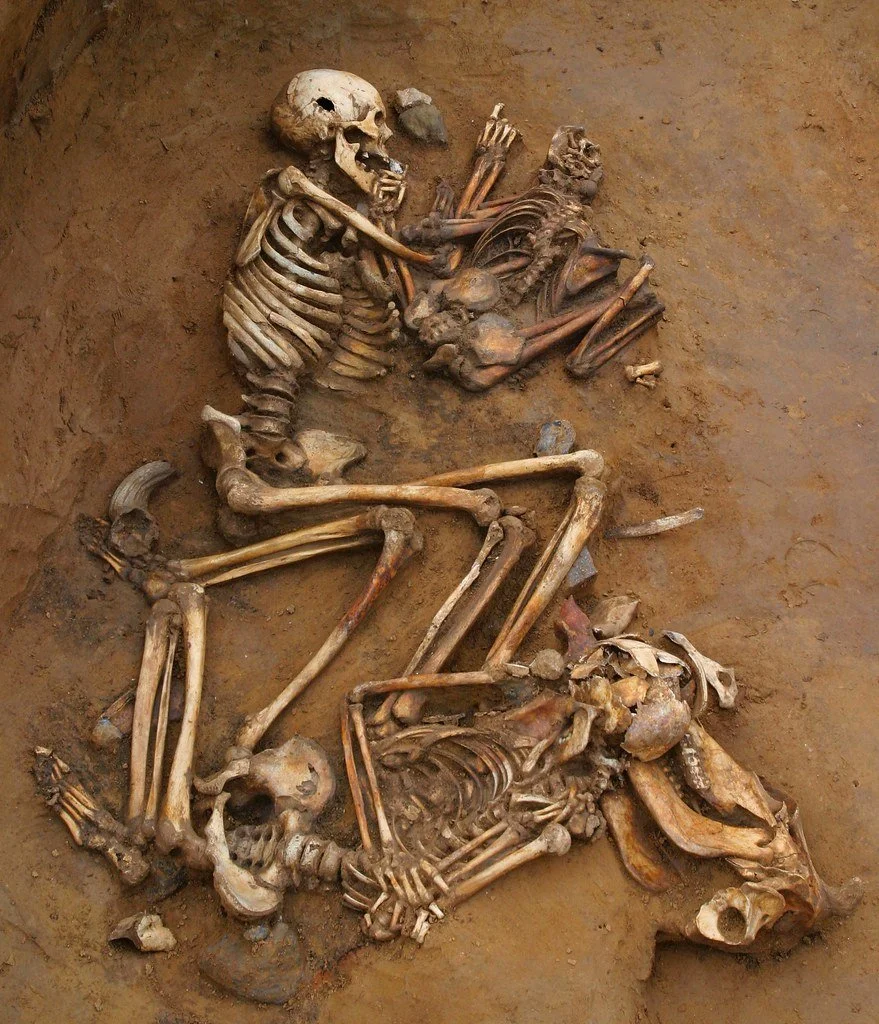
Genomics of kinship between individuals in the Túcume archaeological site, Peru
Some huacas (ancient buildings) were used to bury important people in the ancient times, and these individuals were found buried with other people. In this project, elucidate their inter-relationships via paleo-genomic analysis. There is also the possibility to explore ancestry and issues of health via the detection of ancient pathogens in the human remains.
-

Genetic insights into pre-Incan animal domestication
Remains from guinea pigs, other rodents, camelids, and dogs have been recovered from the Túcume archaeological site, Peru. If the samples correspond to living domesticated animal species that currently live in the region, we can gain knowledge about the origin and nature of their domestications.
-

Genetic identification of pre-Incan ethnobotanical remains
In pre-Columbian Peru, seeds from plants in the genus Nectandra (called amala in Spanish), were used as a muscle relaxant. In the Sican culture, the seeds are thought to have been used as drugs to incapacitate human victims prior to being sacrificed. Now it is possible to buy these seeds in local markets. You can explore this topic by using paleo-genomics to test whether the ancient seeds recovered from archaeological sites match Nectandra species sold in markets today.
-

Paleo-genomics of the Peruvian hairless dog
What is the evolutionary history of the Peruvian hairless dog? By sequencing these ancient dog remains from a burial in Túcume archaeological site, Peru, you can contribute to the knowledge of the evolutionary history of pre-Incan dogs. Specifically, you can determine whether there is a good match with modern dogs of this breed, and then investigate where the breed first appeared and how it evolved into the modern breed.
-

Paleo-genetics of plant domestication in pre-Incan Peru
Samples from diverse food plants such as corn, Cucurbita, Capsicum, Prosopis, avocado, cane, peanut, and Annona (pawpaw) have been excavated from the Túcume archaeological site, Peru. Use paleo-genomics to answer questions about the use and long-distance trade of these food plants within pre-Incan Peru, to study their evolution over time, as well as to probe the origins and nature of their domestications.
-

Genomic investigation of pre-Incan feather trade
A large number of feathers recovered from within the huacas at Túcume archaeological site, Peru. Using paleo-genomics, you can identify these materials to particular bird species, and then use their present-day distributions to reconstruct Peru’s pre-Incan trade routes, learn about ancient animal husbandry, or hypothesize about temporal changes in the birds’ distributions.
-

Genomic insights into land snails inhabiting desert lomas
The lomas are geographic formations and unique seasonal ecosystems in the coastal deserts of Peru and Chile, where the main source of humidity is fog during the winter months. Land snails of the genus Bostryx and Scutalus are well adapted to these harsh conditions, including a period of dormancy in the dry season. You can investigate these snails’ special adaptations to the lomas using a genomic approach.
-

Genomic signatures of high-altitude adaptation in Andean fish
Some Andean catfish of the genus Astroblepus are able to thrive at altitudes up to 3500 meters above sea level. And they can climb up waterfalls using specialized adaptations in their mouths and pelvic fins! You can investigate the genomic basis of these adaptations using genomic sequencing data.
-

Biological soil crust metagenomics in the Peruvian desert
A biological soil crust (BSC) exists in the Peruvian desert, one of the driest ecosystems in the world. This BSC community — composed of mosses, lichens, microfungi and cyanobacteria — is vital for seasonal vegetation and desert fauna because they regulate the availability of water and nutrients by acting as an ecosystem engineer. What environmental factors can influence the taxonomic and functional diversity of these BSC communities in these extreme sites? You can investigate this question using metagenomic methods.
Hosted by Universidad Icesi (Colombia)
-

Diversity of root-associated soil microbes in tropical dry forests
Explore how Colombia’s tropical dry forest plants and their soil microbes respond to drought. Use metagenomic analysis to identify the soil microbial communities (bacteria and fungi) that associated with each plant species and determine which root symbionts and pathogens influence the success of these plants during environmental stress.
-

Composition of plant root-associated soil microbes in a high-mountain tropical ecosystem
In Colombia’s high-mountain páramos, the plant Pernettya prostata (Ericaceae) has evolved with specialized, but largely unknown, root-associated fungi and bacteria. These microbes increase in diversity at high altitudes and may enable the plant to thrive by, for example, assisting in the capture of soil nutrients and reducing soil pH. Use metagenomics to identify the root-associated microbes and to investigate their interactions with the host plant.
Hosted by Universidad del Valle (Colombia)
-

Interactions between insects and Araceae plants in Colombia's tropical forests
Colombia’s Araceae plants are characterized big inflorescences that provide resources and shelter to an amazing diversity of insects. Each plant species has its own set of up to 70 species of interacting insects. Study these interactions in tropical forests combining fragrance analysis, ecological surveys, and genomic sequencing data.
Project host: Alejandro Zuluaga Trochez. -

Aroid plant phylogenetics and genomics
The Araceae (aroids) are a family of monocot flowering plants in which flowers are borne on a type of inflorescence called a spadix, usually accompanied by a leaf-like bract. Use genomic sequencing and analysis to study the evolution of flower and leaf in the Neotropical aroid plants.
Project host: Alejandro Zuluaga Trochez.
Hosted by Universidad del Rosario (Colombia)
-

The influence of root-associated fungi in tropical biogeochemical cycling
Combine -omics and soil analysis to answer important questions about, for example, the influence of mycorrhizal fungi in nitrogen and phosphorus acquisition in tropical plants, or how mycorrhizae mediate carbon accumulation in tropical soils.
Hosted by University of Oslo (Norway)
-

Evolutionary history of Amazonian fish
The Amazon River is the largest freshwater river in the world. In this project we will work on phylogenomics and population genomics of charismatic Amazonian fish. Projects may focus on groups such as piranhas, electric fish, etc., to understand their evolutionary history. Collaborator: Jonathan Ready (Federal University of Pará, Brazil).
Project host: José Cerca. -

Adaptive radiation in the Galápagos lava lizards
The Galápagos Islands need no introduction. One might be surprised by how unexplored some taxa are. In this project we will use whole-genome resequencing of Galápagos lava lizards in order to determine whether they are an adaptive radiation. Collaborator: Juan Manuel Guayasamil (USFQ, Ecuador).
Project host: José Cerca.
Hosted by NTNU (Norway)
-

Comparative transcriptomics of Galápagos daisy trees
During the past 4 million years, the endemic Galápagos daisy trees rapidly diversified from a weedy ancestor into the genus Scalesia, which contains at least 15 species with spectacularly different life history traits and variation in the leaf size/shape. While Darwin’s finches have become a model of adaptive radiation in the Galápagos, these plants have received remarkably little attention despite their even more dramatic radiation into diverse ecological niches. In this project, investigate the role of the gene expression in driving Scalesia plants’ adaptive radiation.
Project host: Michael Martin. -

Paleo-genomics of potato domestication
Despite the potato’s status as one of the world’s most important crops, surprisingly little is known about the history and process of its domestication. In this project, utilize ultra-rare genetic data from archaeological specimens of ancient potatoes to reveal how the use of potatoes by ancient South American peoples led to today’s diverse potato varieties.
Project host: Michael Martin. -

Urban evolution of Psittacara parrots from Lima (Peru)
Psittacara (Psittacidae) is one of the most diverse and charismatic groups of parrots from the Neotropics. Four out of their twelve recognized species are distributed in the Peruvian jungle. Because these birds have been illegally traded for many decades, in Peruvian cities like Lima it is common to find Psittacara populations founded by trafficked individuals that escaped. In this project, you can test for inter-specific gene flow between Lima’s Psittacara parrots using nearly 100 samples from the Natural History Museum of San Marcos.
Project host: Jaime G. Morin Lagos.
